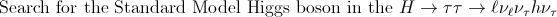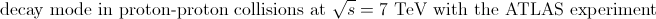| |
|
| Research Interests: |
- Mathematical modelling of receptor tyrosine kinase crosstalk mechanisms in
cancer.
- Linking model-derived features to cell fate via machine
learning
- Transcription factor networks for developmental
timing in Zebrafish, RNAseq analysis
- Approximation of prediction
and validation bands via integration methods(Download of analytical
ABC example)
- Optimization techniques, sensitivity computation
- Co-Developer of open-source software Data2Dynamics
|
|
|
|
| Publications: |
| H. Hass, F. Kipkeew, A. Gauhar, E. Bouché, P. May, J. Timmer, H.H. Bock |
| Mathematical model of early Reelin-induced Src family kinase-mediated signaling. |
| PLoS ONE (2017) 12(10): e0186927 |
|
| H. Hass, K. Masson, S. Wohlgemuth, V. Paragas, J.E. Allen, M. Sevecka, E. Pace, J.
Timmer, J. Stelling, G. MacBeath, B. Schoeberl, A. Raue |
| Predicting ligand-dependent tumors from multi-dimensional signaling features. |
| npj Systems Biology and Applications (2017)
3(1): 27 |
|
| D. Kurzhunov, R. Borowiak, H. Hass, P. Wagner, A. Krafft, J. Timmer M. Bock |
| In vivo quantification of oxygen metabolic rates in the human brain with dynamic 17 O MRI: profile likelihood analysis. |
| Magnetic Resonance in Medicine (2016)
78(3): 1157-1167 |
|
| T. Maiwald, H. Hass, B. Steiert (shared), J. Vanlier, R. Engesser, A. Raue, F. Kipkeew, H.H. Bock, D. Kaschek, C. Kreutz, J. Timmer |
| Driving the model to its limit: profile likelihood based model reduction. |
| PloS ONE (2016) 11(9): e0162366 |
|
| R. Merkle, B. Steiert, F. Salopiata, S. Depner, A. Raue, N. Iwamoto, M. Schelker, H. Hass, M. Wäsch, M. Böhm, O. Mäcke, D.B. Lipka, C. Plass, W.D. Lehmann, C. Kreutz, J. Timmer, M. Schilling, U. Klingmüller |
| Identification of cell type-specific differences in erythropoietin receptor signaling in primary erythroid and lung cancer cells. |
| PloS
Comp. Biology (2016) 12(8): e1005049 |
|
| H. Hass, C. Kreutz, J. Timmer, D. Kaschek |
| Fast integration-based prediction bands for ordinary differential equation models |
| Bioinformatics
(2016) 32(8): 1204-1210 |
|
| A. Raue, B. Steiert, M. Schelker, C. Kreutz,
T. Maiwald, H. Hass, J. Vanlier, C. Tönsing, L. Adlung,
R. Engesser, W. Mader, T. Heinemann, J. Hasenauer,
M. Schilling, T. Höfer, E. Klipp, F. Theis, U. Klingmüller,
B. Schoberl, J. Timmer |
| Data2Dynamics: a modeling environment tailored to parameter estimation in dynamical systems |
| Bioinformatics (2015) 31(21): 3558-3560. |
|
|
| Invited Talks: |
| 07/2018 | Combining mechanistic modeling of receptor crosstalk with machine learning to predict tumor growth |
7th Conference on Systems Biology of Mammalian Cells, SBMC2018 |
| 09/2017 | Profile-likelihood based model reduction |
Workshop: ODE Modelling in Systems Biology |
| 07/2017 | Predicting tumor growth and ligand dependence from mRNA by combining mechanistic
modeling with machine learning |
Conference of Systems Biology of Human Disease: SBHD2017 |
| 02/2017 | Mechanism-based learning connects response to growth factors with their expression in
tumors |
Bioinformatics Club Freiburg |
|
|
|
| Awards: |
|
MTZ-Award 2018 für herausragende Dissertationsarbeiten auf dem
Gebiet der medizinisch orientierten Systembiologie |
| Alumni Award
2014 from the University of Freiburg |
| 10/08 - 04/13: Scholarship awarded from Friedrich-Ebert-Stiftung |
| 01/08 - 04/13: Scholarship
awarded from e-fellows.net |
|
|
| |
| Curriculum Vitae:
| | |
| detailed CV |
| Since 01/2018 | Postdoctoral Researcher in the group of
Prof. Dr. Jens Timmer,
Albert-Ludwigs-Universität Freiburg |
| 02/2016 - 09/2016 | Intern at Merrimack Pharmaceuticals, Boston, MA, USA |
| 01/2014 - 12/2017 | PhD student in the group of Prof. Dr. Jens Timmer,
Albert-Ludwigs-Universität Freiburg
Quantifying cell biology: Mechanistic dynamic modeling of
receptor crosstalk |
| 10/2005 - 04/2013 | Student of physics at
Albert-Ludwigs-Universität Freiburg
Diploma Thesis
(advisor Prof. Dr. Karl Jakobs):
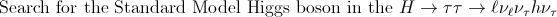 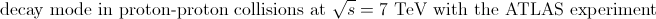
|
| 10/2007 - 07/2011 | Bachelor study of economics at
Albert-Ludwigs-Universität Freiburg |
Datenschutz
1. Verantwortlich im datenschutzrechtlichen Sinne:
Helge Hass
Habsburger Strasse 49
79098 Freiburg
hhass at physik.uni-freiburg.de (replace " at " by @)
2. Datenschutzbeauftragter
Albert-Ludwigs-Universität Freiburg
Datenschutzbeauftragter
datenschutz at uni-freiburg.de (replace " at " by @)
3. Ihre Rechte
- Sie haben das Recht, von der Universität Auskunft über die zu Ihrer Person gespeicherten Daten zu erhalten und/oder unrichtig gespeicherte Daten berichtigen zu lassen.
- Sie haben darüber hinaus das Recht auf Löschung oder auf Einschränkung der Verarbeitung oder ein Widerspruchsrecht gegen die Verarbeitung.
- Sie haben das Recht die Sie betreffenden personenbezogenen Daten, die Sie uns bereitgestellt haben, in einem strukturierten, gängigen und maschinenlesbaren Format zu erhalten, und Sie habe das Recht, diese Daten einem anderen Verantwortlichen ohne Behinderung durch uns zu übermitteln.
- Sie haben das Recht auf Beschwerde bei der Aufsichtsbehörde, wenn Sie der Ansicht sind, dass die Verarbeitung der Sie betreffenden personenbezogenen Daten gegen die Rechtvorschriften verstösst.
Die zuständige Aufsichtsbehörde ist der Landesbeauftragte für den Datenschutz und die Informationsfreiheit Baden-Württemberg.
4. Zugriff auf die Webseite
Die Webseite verwendet keine Cookies oder sonstige Tools, die personenbezogene Daten abspeichern oder analysieren.
Dennoch können serverseitig automatisch Informationen allgemeiner Natur erfasst werden. Diese Informationen (Server-Logfiles) beinhalten etwa die Art des Webbrowsers, das verwendete Betriebssystem, den Domainnamen Ihres Internet-Service-Providers und ähnliches. Hierbei handelt es sich ausschließlich um Informationen, welche keine Rückschlüsse auf Ihre Person zulassen.
Diese Informationen sind technisch notwendig, um von Ihnen angeforderte Inhalte von Webseiten korrekt auszuliefern und fallen bei Nutzung des Internets zwingend an.
| |